
Poor prognosis and aggressive features of primary adrenal DLBCL linked to PD-L1 gene alterations
Primary adrenal (PA) lymphoma is extremely rare, accounting for less than 1% of all non-Hodgkin lymphomas, yet is very aggressive and has a poor prognosis, with a general survival time of less than one year. In most cases, PA lymphomas are derived from B cells, with diffuse large B cell lymphoma (DLBCL) being the most common phenotype. The genetic features that contribute to the development of PA-DLBCL have yet to be fully described, but previous research has suggested that alterations in the gene that codes for programmed death ligand 1 (PD-L1) may contribute to the development of DLBCL in specific primary sites, including the testis, mediastinum, central nervous system, and adrenal gland. In this study, the authors investigated the frequency of PD-L1 gene alterations in 18 cases of PA-DLBCL and looked for an association between PD-L1 gene alterations and clinicopathologic variables, including prognosis.
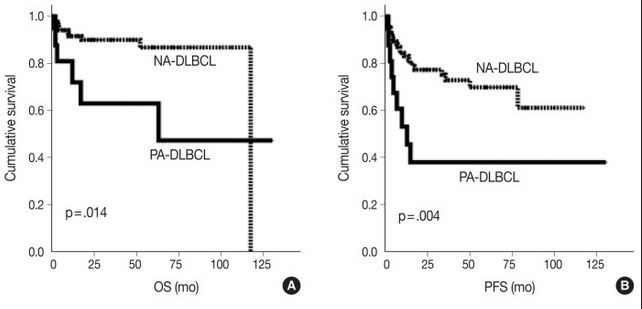
For this study, the authors defined PA-DLBCL as “de novo DLBCL with one or more adrenal gland mass-forming and/or hypermetabolic lesion(s) as the most dominant lesion”. A total of 18 cases were retrospectively retrieved from the archives of pathology records at Seoul National University Bundang Hospital to represent the PA-DLBCL group. Ninety systemic cases of DLBCL, NOS diagnosed at the same hospital during the same time period were used as a non-adrenal (NA)-DLBCL reference group. In comparison to the NA-DLBCL control group, patients in the PA-DLBCL group were more likely to have a high IPI score (3-5; p = 0.007), poor ECOG performance score (≥2; p = 0.003), expression of MUM1 (p = 0.047) and elevated LDH (p = 0.035), with trends for high frequency of B symptoms (p = 0.084) and bulky disease (p = 0.084), despite both groups showing a similar distribution of high-stage disease (50% in PA-DLBCL vs. 53% in NA-DLBCL). The PA-DLBCL group also showed inferior overall survival (OS) and progression-free survival (PFS) when compared to the NA-DLBCL group (p = 0.014 and p = 0.004, respectively). Thus, patients in the PA-DLBCL group harbored more aggressive clinicopathologic features and a poorer prognosis compared to patients in the NA-DLBCL group, even with a similar disease stage.

Next, fluorescent in situ hybridization (FISH) was used to investigate the frequency of genetic alterations of the gene that encodes PD-L1 (CD274). The authors used a break-apart FISH probe to detect translocations and a PD-L1 gene/chromosome 9 probe set to detect copy number gain and amplification. For each case, signals were counted in more than 200 cells with non-overlapping nuclei. Separation of signals in more than 15% of cells was interpreted as the presence of translocation, and PD-L1 gene/chromosome 9 ratios >4 was considered amplification. The PA-DLBCL group had a significantly higher frequency of PD-L1 gene alterations than the NA-DLBCL group (39% vs. 6%, respectively; p = 0.001), with translocations in 22% vs. 3% of cases (p = 0.016) and amplifications in 17% vs. 2% of cases (p = 0.034). This suggests that PA-DLBCL may be a distinct entity with unique genetic alterations.

Finally, the authors analyzed associations between the PD-L1 gene alterations and clinicopathological parameters and prognosis within the PA-DLBCL group. Among patients with PA-DLBCL, those with alterations in the gene for PD-L1 tended to have elevated serum LDH, presence of B symptoms, and less bulky disease, although these trends were nonsignificant (p = 0.119, p = 0.145, and p = 0.119, respectively). When survival was analyzed within the PA-DLBCL group, PD-L1 gene alterations as a whole did not significantly affect OS or PFS. However, further analysis revealed that, while PD-L1 gene amplification was not associated with prognosis, PD-L1 translocations contributed to a significantly inferior OS (p<0.001) and PFS (p = 0.012). Though the aggressive clinicopathologic nature of PA-DLBCL cannot be fully explained by PD-L1 gene alteration alone, as PD-L1 gene alteration was only identified in 39% of cases, this study demonstrates that PA-DLBCL has aggressive features with a high rate of PD-L1 gene alteration and poor prognosis. Further research is needed to elucidate the role of PD-L1 signaling in the pathogenesis of PA-DLBCL and the association between PD-L1 expression and aggressive disease.
