
Proteomic differences between squamous cell carcinomas from rare and common primary sites
Squamous cell carcinoma (SCC), which develops in the top layer of the epithelium, and adenocarcinoma (AC), which develops in glandular epithelium cells, are two of the primary histological subtypes of carcinoma. Common primary sites for squamous cell carcinoma include the oral cavity, throat, skin, esophagus, lung, and cervix, among others; common primary sites for adenocarcinomas include the thyroid, lung, breast, pancreas, and colorectum, among others. While better prognosis and treatment options based on the molecular mechanisms of tumor formation and progression in AC are emerging, these options remain limited for many SCCs. Though SCCs share histological characteristics, and several studies have identified recurrent mutations among SCCs, these have shown limited value for predicting prognosis or optimal therapy and have been unable to reliably distinguish site of origin for metastatic SCC. This study takes a proteomics-based approach to characterize SCC, provides functional insights that may assist with SCC diagnosis and prognosis, and proposes avenues to explore for future SCC treatment targets.
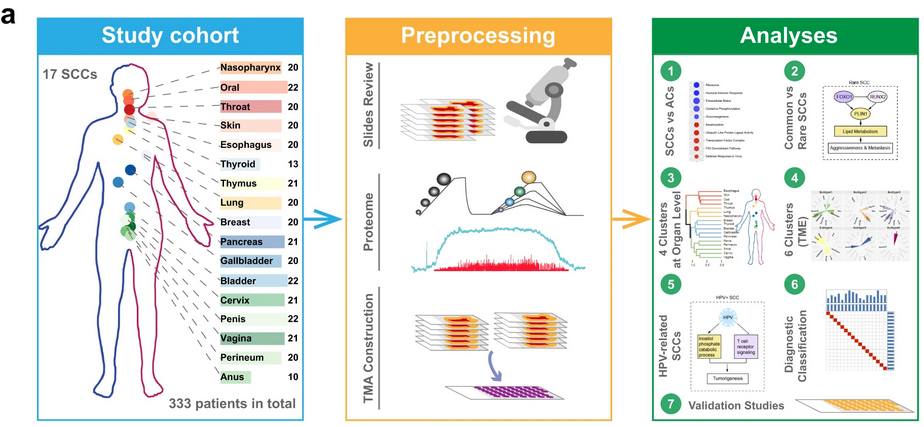
Formalin-fixed paraffin-embedded tissue samples from 333 treatment-naïve patients with SCC who underwent surgical resection at Zhongshan Hospital, Fudan University between May 2001 and December 2019 were randomly selected from archive to comprise the pan-SCC cohort. Proteomic analysis was conducted using liquid chromatography-tandem mass spectrometry (LC-MS/MS) and, in total, 14,840 protein groups were quantified in the pan-SCC cohort, with an average of 8120 protein groups per sample. As expected, high protein levels of the SCC diagnostic markers KRT5 and TP63 were found across samples from all 17 primary sites. Ubiquitous decreases in protein levels were measured for commonly mutated tumor suppressors, including TP53, PTEN, and FGFR3. To examine functional differences between SCCs and ACs, 69 randomly selected archived samples were collected from 69 treatment-naïve adenocarcinoma patients that underwent surgery between July 2017 and August 2019. Overall, differentially expressed proteins between SCCs and ACs were in line with the properties of the epithelial tissues from which they originated. Gene set enrichment analysis revealed that the pathways enriched in SCCs included those involved in keratinization, ubiquitin-like protein ligase activity, transcription factor complex, p53 downstream pathway, and defense response to virus. Pathways enriched in ACs included those involved in the ribosome, humoral immune response, extra-cellular matrix (ECM), oxidative phosphorylation, and gluconeogenesis.
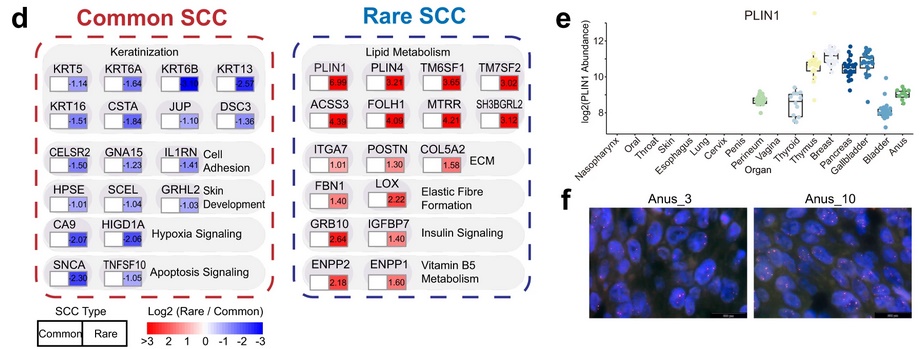
Within the pan-SCC sample cohort, ten common SCC sites (nasopharynx, oral cavity, throat, skin, esophagus, lung, cervix, penis, vagina, and perineum) and seven rare SCC sites (thyroid, thymus, breast, pancreas, gallbladder, bladder, and anus) were represented. As proteins such as YAP1, AKT3, and SOX2 had shown differential expression levels between SCCs in the original analysis, the authors decided to investigate how different pathological parameters, including common vs. rare primary sites, influenced the SCC proteome. Principal component analysis revealed a significant difference in protein levels between common SCCs and rare SCCs for 938 proteins (500 proteins upregulated in common SCCs and 438 proteins upregulated in rare SCCs). Pathway enrichment analysis revealed significant enrichment of keratinization, cell adhesion, skin development, and apoptosis signaling pathways among common SCCs. Pathways enriched among rare SCCs included those involved in lipid metabolism, ECM organization, elastic fiber formation, insulin signaling, and vitamin B5 metabolism. Specifically, levels of Perilipin 1 (PLIN1), involved in lipid metabolism, were very high in rare SCCs and not detected in most common SCCs. Fluorescent in situ hybridization revealed PLIN1 gene amplification in 3/10 anal SCCs, suggesting one plausible reason for these elevated levels. The transcription factors RUNX2 and FOXO1 were also uniquely overexpressed in rare SCCs. The authors propose that lipid metabolism may reinforce the malignancy of rare SCCs, and suggest that staining for PLIN1, RUNX2, and FOXO1 could be used in the diagnosis of rare SCCs.
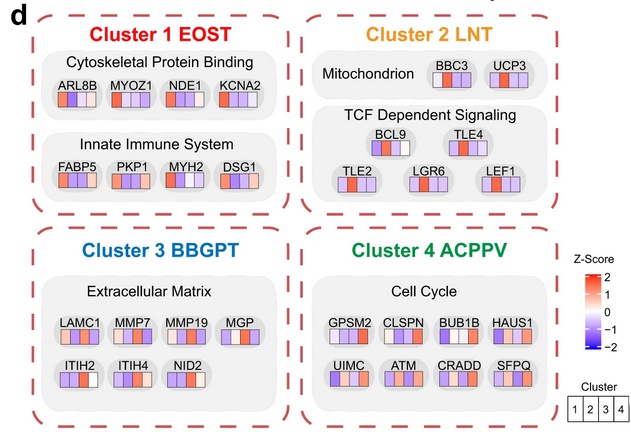
To identify proteomic features and relationships of possible biological and clinical relevance, the authors used hierarchical clustering to group the SCCs from all 17 primary sites into subgroups. Four proteomic subtypes emerged from this analysis. Cluster 1 contained esophagus, skin, oral, and throat SCCs and featured significantly enriched proteins associated with cytoskeletal binding and the innate immune system. Cluster 2 contained lung, nasopharynx, and thymus SCCs and featured significantly enriched proteins associated with the mitochondrion and TCF-dependent signaling. Cluster 3 contained 5 of the 7 rare SCCs including bladder, breast, gallbladder, pancreas, and thyroid; the proteins enriched in this cluster included those associated with the ECM. Cluster 4 contained all anogenital SCCs, including anus, cervix, penis, perineum, and vagina; the proteins enriched in this cluster were involved in the cell cycle. These differences could indicate possible differential SCC initiation, though much more research would be needed to conclude. Cluster analysis was also conducted based on immune features of the tumor microenvironment (TME); this analysis identified 6 SCC subtypes with different TME features and prognosis. As many new cancer therapeutics target features of the TME, the authors propose drug targets to explore for each subgroup, including sodium channels, potassium voltage-gated channels, voltage-dependent calcium channels, and solute carrier family transporters. Though further research is required, the proteomics data presented here provide a foundation on which to build future clinical and translational SCC studies.
