Oncogenic lncRNA PCAT2 linked to chromosome instability in cancer
Chromosome breaks and translocations are often critical factors in tumorigenesis and cancer progression. One primary contributor to these types of chromosomal instability is the mis-regulation of histone proteins. Histones are responsible for packaging the human genome into chromatin and can be divided into five main families: H1/H5, H2A, H2B, H3, and H4. The histone H3 has several variants, including centromeric protein A (CENP-A), which is essential for mitosis and is generally only enriched in centromere-specific DNA regions. Inefficient loading of CENP-A during the early G1 phase results in downstream accumulation of mitotic defects. CENP-A is overexpressed in cancer cells, and overexpression of CENP-A is linked with a poor clinical prognosis. CENP-A’s correlation with disease severity has been attributed to its mis-localization outside of the centromere to chromosome arms and telomeres, sensitizing those areas to chromosome instability and subsequent damage. However, the mechanism by which CENP-A gets deposited at these genomic regions outside the centromere has yet to be established. In this study, the authors examined CENP-A mis-localization to 8q24, a chromosomal region that has a large CENP-A domain in patient-derived colon cancer cells and contains the cMYC gene, which is amplified or translocated in numerous cancers. Other than cMYC, the 8q24 region encodes five long noncoding RNAs (lncRNAs) that have been clinically associated with cell proliferation, metastasis, and therapeutic resistance to treatment in cancer. Thus, the authors looked for a relationship between the transcriptional activity of oncogenic lncRNAs arising from a specific locus and the mis-localization of histone variants to that locus.
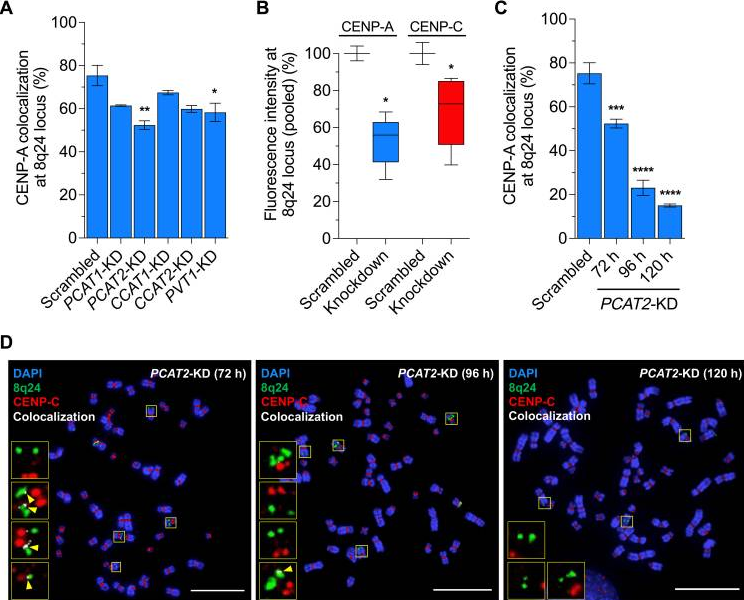
First, real time qPCR was used to determine relative expression levels of the 8q24 lncRNAs (PVT1, PCAT1, PCAT2, CCAT1, and CCAT2) in the SW480 metastatic colon cancer cell line. CCAT1 was most highly expressed, followed by PCAT2, PCAT1, PVT1, and CCAT2. Antisense oligo (ASO)-mediated knockdown of the five lncRNAs was used to examine the consequences of the loss of these lncRNAs on ectopic CENP-A distribution. Colocalization of CENP-A and CENP-C, which localizes to ectopic sites along with CENP-A in cancer cells and was used as a proxy for CENP-A staining, to the 8q24 locus was quantified using IF/FISH. The knockdown of the five lncRNAs resulted in a depletion of CENP-A and CENP-C at the 8q24 locus (p < 0.05), with the most pronounced decrease seen with the PCAT2 knockdown (p < 0.01). At 120 hours after depletion, CENP-A and CENP-C occupancy at the 8q24 locus decreased to 15% and 23% of the control, respectively. In addition, the lncRNA knockdown depleted CENP-C only at the 8q24 locus and not at two other loci (2q21 and 10p15), suggesting that the lncRNAs at the 8q24 locus function in cis to recruit CENP-A/CENP-C to the locus from which they transcribe.
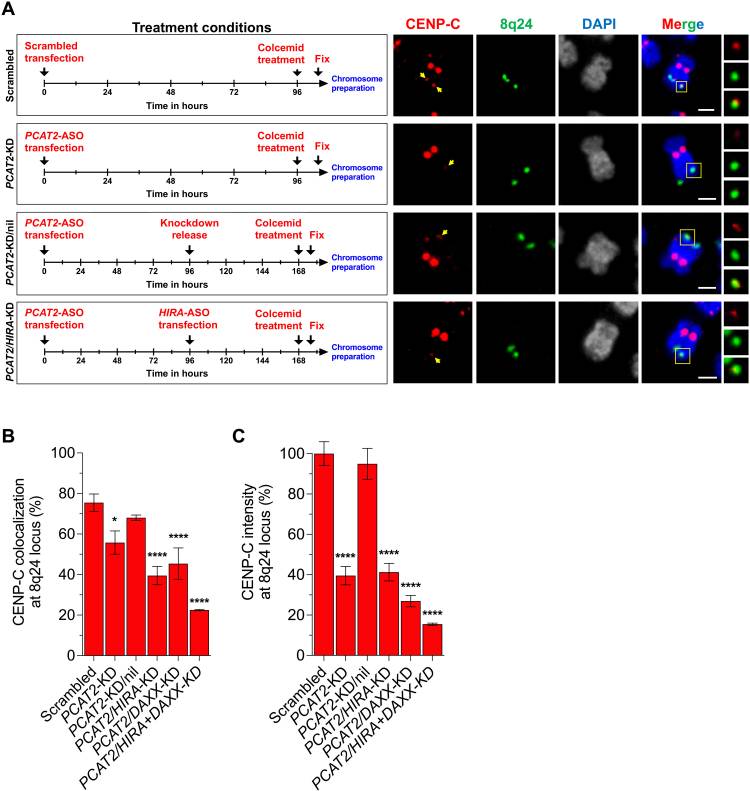
Normally, the chaperone protein HJURP deposits CENP-A at centromeres. However, recent work has reported that overexpression of CENP-A results in an association between CENP-A and the H3.3 chaperones HIRA and DAXX, and that this association is involved in CENP-A deposition at ectopic sites. Thus, the authors next investigated whether PCAT2 interacts with CENP-A via the H3.3 chaperones HIRA and DAXX through sequential ASO-knockdowns. IF/FISH was used to measure CENP-C levels at the 8q24 locus for each treatment condition. As observed in the earlier experiment, PCAT2 knockdown reduced the amount of CENP-C deposited at the 8q24 locus, but the sequential knockdowns of PCAT2, HIRA, and DAXX resulted in even lower CENP-C levels (p<0.0001), and resulted in chromosome deformities and a significant number of nonviable cells. When released from the PCAT2 knockdown, the amount of CENP-C at the 8q24 locus remained low without HIRA or DAXX. The authors interpret these results to mean that PCAT2 cannot cause the mis-localization of CENP-A without the cooperation of the H3.3 chaperones HIRA or DAXX.
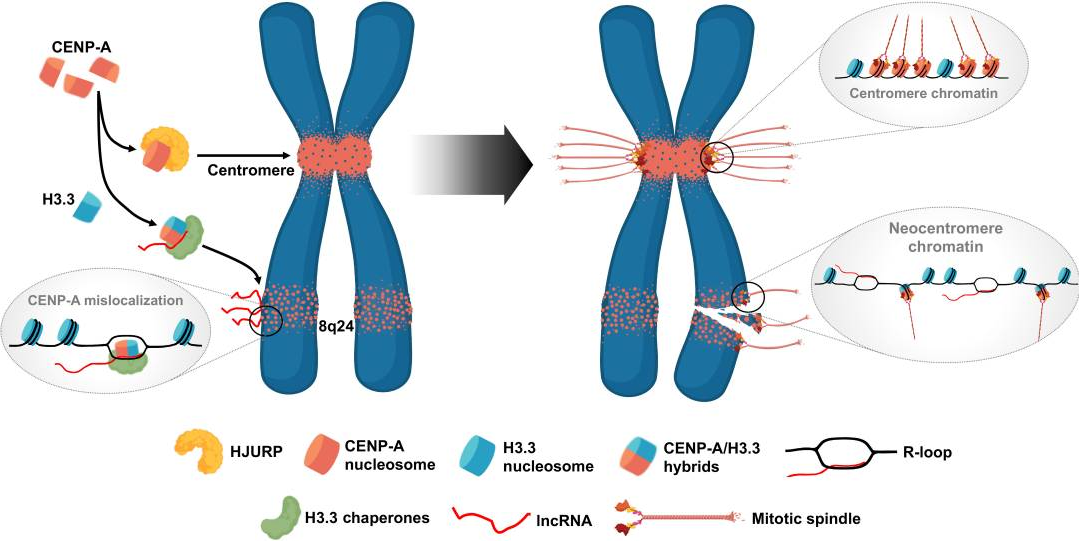
Finally, the authors investigated if introducing the lncRNA PCAT2 gene to a naïve chromosome locus could result in the mis-localization of CENP-A to that locus. A transgene cassette carrying the PCAT2 gene was inserted at the 4q31 locus that lacks lncRNA genes and ectopic CENP-A but contains a known fragile site. The cells that received the transgene PCAT2 had significantly higher levels of CENP-C at the 4q31 locus than wild type cells that did not receive the TransPCAT2 cassette. In fact, at week 24, the 4q31 locus showed a fourfold increase in CENP-C compared to wild type, and even outcompeted the native 8q24 locus. This shows that the oncogenic lncRNA PCAT2 is able to both recruit and retain CENP-A to any region of the genome. Selective knockdown of the TransPCAT2 RNA reduced the CENP-C levels at the 4q31 locus but not at the 8q24 locus, confirming that PCAT2 works in cis, not in trans. Taken together, the authors propose a working model by which lncRNA PCAT2 can mimic the function of centromeric lncRNAs in the context of a CENP-A:HIRA/DAXX complex, causing mis-localization of CENP-A to chromosome arms and telomeres, but using pathways that mimic normal CENP-A deposition at centromeres. Thus, further research should focus on finding small-molecule inhibitors that could exploit this interaction as a therapeutic target for several human cancers.