Point mutation causes dramatic shift in HIV-1 integration sites
Retroviruses, such as the human immunodeficiency virus (HIV-1), insert a copy of their genome into the host cell genome during a process called integration. This allows retroviruses to persist indefinitely in the infected cell as a provirus. During integration, the virally encoded integrase (IN) protein binds to various host factors that likely act as a tether between the viral IN protein and the host chromatin at the integration site. These integration sites are not randomly distributed throughout the genome; for example, HIV-1 IN displays a significant preference for integrating into actively transcribed regions of the human chromosome. In this study, a point mutation at the K258 acetylation site in the C-terminal domain of HIV-1 IN resulted in a significant and dramatic shift in the distribution of integration sites to alphoid repeat sequences within centromeres. As centromeric HIV-1 integrations have been linked to viral latency and control, understanding the redirection of integrations to the centromere may provide important insight about long term control in patients.
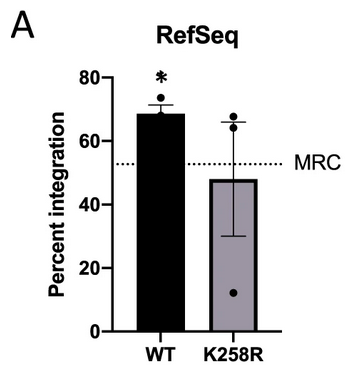
The authors mutated each of four major acetylation sites in the C-terminal domain of HIV-1 IN (K258, K264, K266, and K273) by changing lysine residues to charge-conservative arginines. HIV-1 reporter constructs were created and packaged with WT IN or a mutant IN into virion particles that were used to infect HeLa cells. Infected cells were collected at 48 hours post-infection and all viral DNA intermediates and viral mRNA levels were quantified using qPCR. The K258R point mutation in IN was shown to significantly decrease the amount of reverse transcription products (p <0.01) and levels of integration as assessed by proviral DNA formed (p < 0.001). High-throughput DNA sequencing of infected cells was used to find viral-host integration junctions, and these junctions were mapped to human sequences using Bowtie2. Three of the four mutant INs, as well as the combinatorial quadruple acetylation mutant IN, showed little deviation from the WT integration distribution. However, the K258R mutant proviral DNA was significantly less likely to be located in known genes (p<0.05).
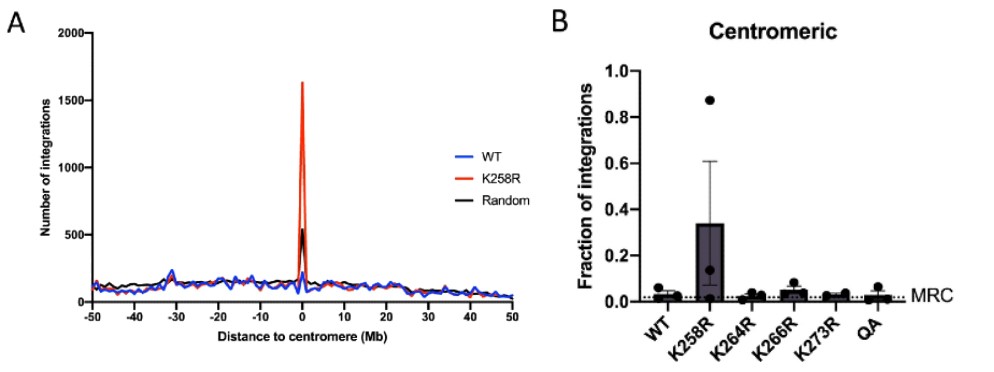
To determine where the K258R mutant IN was being redirected, a custom perl script was used to identify regions of the genome (10kb or less) with five or more viral integrations. A large number of these “hot spot” sites for K258R integration were identified, and they all clustered in centromeres. WT HIV-1 IN, on the other hand, had no detectable integration hot spots located within the centromere. Further analysis revealed that approximately 8% of total K258R proviruses were integrated into centromeres, which represented a tenfold increase in frequency compared to WT (p<0.0001). Of note, the quadruple acetylation mutant did not produce the same phenotype, nor did any of the other mutant INs redirect integration into centromeres as dramatically. Mapping by the RepeatMasker track from the UCSC Genome Browser revealed that the K258R IN mutation causes a specific targeting of integrations into alphoid repeat sequences but not to other classes of repeat sequences. The two preferred spots of integration are at nucleotide position 6 and 69 in the alphoid consensus sequence, though the reason for this preference is unclear.
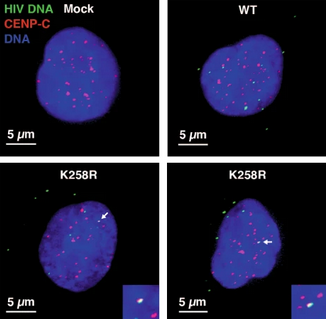
The authors verified the preference for integration into centromeres using other methods, including florescent in situ hybridization (FISH), in which HIV-1 DNA signals detected in the nucleus were counted for the colocalization with the centromere. This assay revealed an average 12-fold increase in association between the K258R proviral DNA and the centromere compared to WT proviral DNA (p<0.05). This confirmed that the K258R mutant IN redirected integration to centromeric regions. While the authors attempted to determine the role of various protein host factors in the retargeting of K258R HIV-1 IN, their data were inconclusive, with no clear basis for the selectivity of integration to centromeres. Further research is needed to elucidate the functional importance of any of these host factors in creating the K258R phenotype of centromeric integration.