Clinical & Research Blog
Clonal heterogeneity and specific chromosome gains proposed as new prognostic biomarkers for HHD-B-ALL
B-cell acute lymphoblastic leukemia (B-ALL), the most common childhood cancer, features a substantial subgroup of patients with chromosomal gains (hyperdiploidy). Patients with a modal chromosome number >50 (high hyperdiploidy; HHD) account for nearly 30% of B-ALL cases and typically have a more favorable prognosis. Although HHD represents an important prognostic factor in childhood B-ALL, the specific chromosome gains that most contribute to a favorable clinical outcome have yet to be established. In addition, research examining the role of clonal heterogeneity in HHD-B-ALL is limited. Whereas up to 20% of HHD-B-ALL patients eventually relapse, improved risk-stratification is crucial to identifying those patients that are at high risk of disease progression and relapse. In this study, the authors used sequential fluorescent in situ hybridization on interphase nuclei (iFISH) and single-cell computational modeling to characterize clonal heterogeneity in HDD-B-ALL. Significantly, they report that low levels of clonal heterogeneity and the rates of specific chromosome gains are robust relapse risk-predictors in HHD-B-ALL and could be used as prognostic biomarkers at disease presentation.

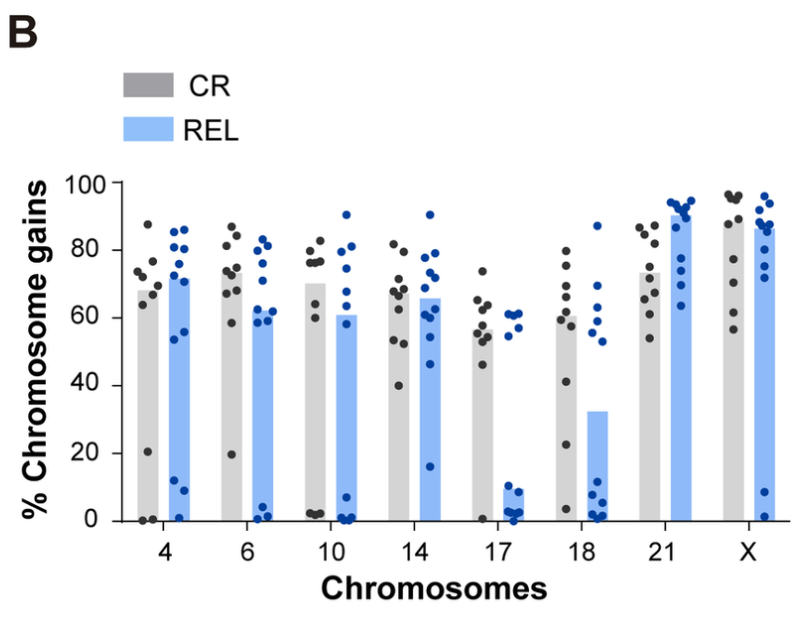
Bone marrow samples from HHD-B-ALL pediatric patients were obtained at disease presentation from collaborating hospitals. Among these diagnosis samples (DX, n=22) were samples from patients who remained alive in complete remission after a minimum 5-year follow up (CR, n=10) and patients who relapsed within this timeframe (REL, n=12). Sequential FISH (seq-iFISH) analyses were performed to assess the presence of the 8 chromosomes typically gained in HHD-B-ALL (chromosomes X, 4, 6, 10, 14, 17, 18 and 21). There was no difference in the total number of chromosomal gains observed in CR versus REL patients. However, there were observed differences in rates of specific chromosomal gains between these two groups. Samples from CR patients had increased rates of gain of chromosomes 17, 18, 6, and 10, whereas samples from REL patients had increased rates of chromosome 21.
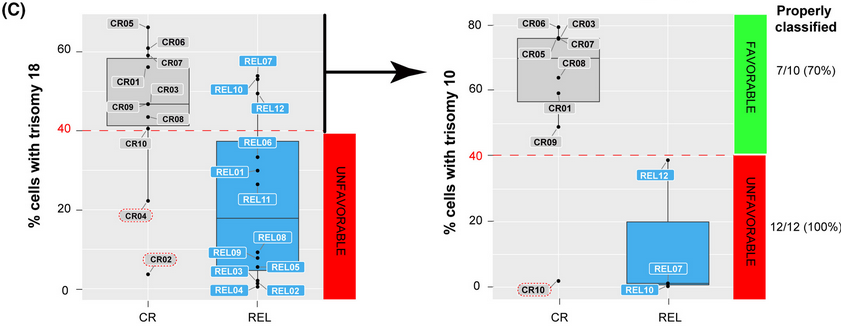
Next, computational modeling using a Random Forest decision tree-based approach examined whether specific combinations of chromosomal gains in DX samples could distinguish patients in CR vs. REL. The most stable and accurate computationally-defined risk predictor was based on chromosomes 18 and 10 trisomy rates. This risk-predictor correctly classified DX patients as CR or REL with 70% and 100% accuracy, respectively. In a blind validation test using a larger independent cohort of HHD-B-ALL samples from 50 patients, this risk-predictor correctly classified 84% (42/50) of patients as either disease-free or relapsed. To verify the prognostic value of their findings, the authors statistically analyzed the effects of age, gender, WBCs, minimal residual disease (MRD) status, treatment protocol and the risk-predictor based on trisomies 18 and 10 on patient outcome in their validation cohort. The risk-predictor based on low levels of trisomies 18 and 10 was the only independent risk factor associated with higher REL rates (p<0.001). Patients with a favorable risk predictor (+18+10>40%) had a higher rate of relapse-free survival than patients with an unfavorable risk predictor (p<0.0001).
Shannon entropy indices were also calculated for each DX sample as a measure of chromosomal instability, which is directly linked to clonal heterogeneity. Significantly higher levels of clonal heterogeneity were observed in CR samples than in REL samples (p=0.036). This association held true in the validation cohort; patients with greater clonal heterogeneity (percentage of major clone; PMC <50%) had a relapse-free survival (RFS) rate of 79.8% beyond five years while patients with lower clonal heterogeneity (PMC >50%) had a RFS rate of 33.3% over the same time period (p=0.0001). In addition, the rates of trisomies 18 and 10 were associated with clonal heterogeneity; as the PMC increased, the rates of trisomies 18 and 10 decreased (p<0.0001). As a result, the authors propose routine FISH testing of HHD-B-ALL patients for low levels of clonal heterogeneity and low levels of trisomies 18 and 10, as these were demonstrated to be robust and independent relapse risk-predictors that would allow for a reliable prognosis at the time of disease presentation.