Clinical & Research Blog
Associations Between CCND1 Amplification in Breast Cancer & Proliferation Status
Breast cancer is a highly heterogeneous disease that differs greatly between patients. As such, there is a need for new biomarker discovery that would allow for individualized diagnoses, treatments, and prognoses for breast cancer patients. In this study, the authors assessed the ability of CCND1 amplification to serve as a prognostic biomarker in breast cancer. CCND1 is located on 11q13 and encodes cyclin D-1 protein. Among other functions, cyclin D-1 activates the cyclin-dependent kinases Cdk4 and Cdk6, which are involved in a signal transduction pathway that results in progression from G1 to S phase of the cell cycle. Cyclin D-1 overexpression is reported in approximately half of breast cancers. Previous studies have noted the frequency of CCND1 amplification to be between 9-15% of breast cancer cases and have measured associations between CCND1 amplification and high proliferation, high histopathological grade, and the Luminal B subtype. The goal of this study was to further investigate associations between CCND1 amplification in primary breast cancer tumors and proliferation status, molecular subtype, and prognosis. The authors also considered the relationship between CCND1 amplification in the primary tumor and CCND1 amplification in corresponding axillary lymph node metastases.
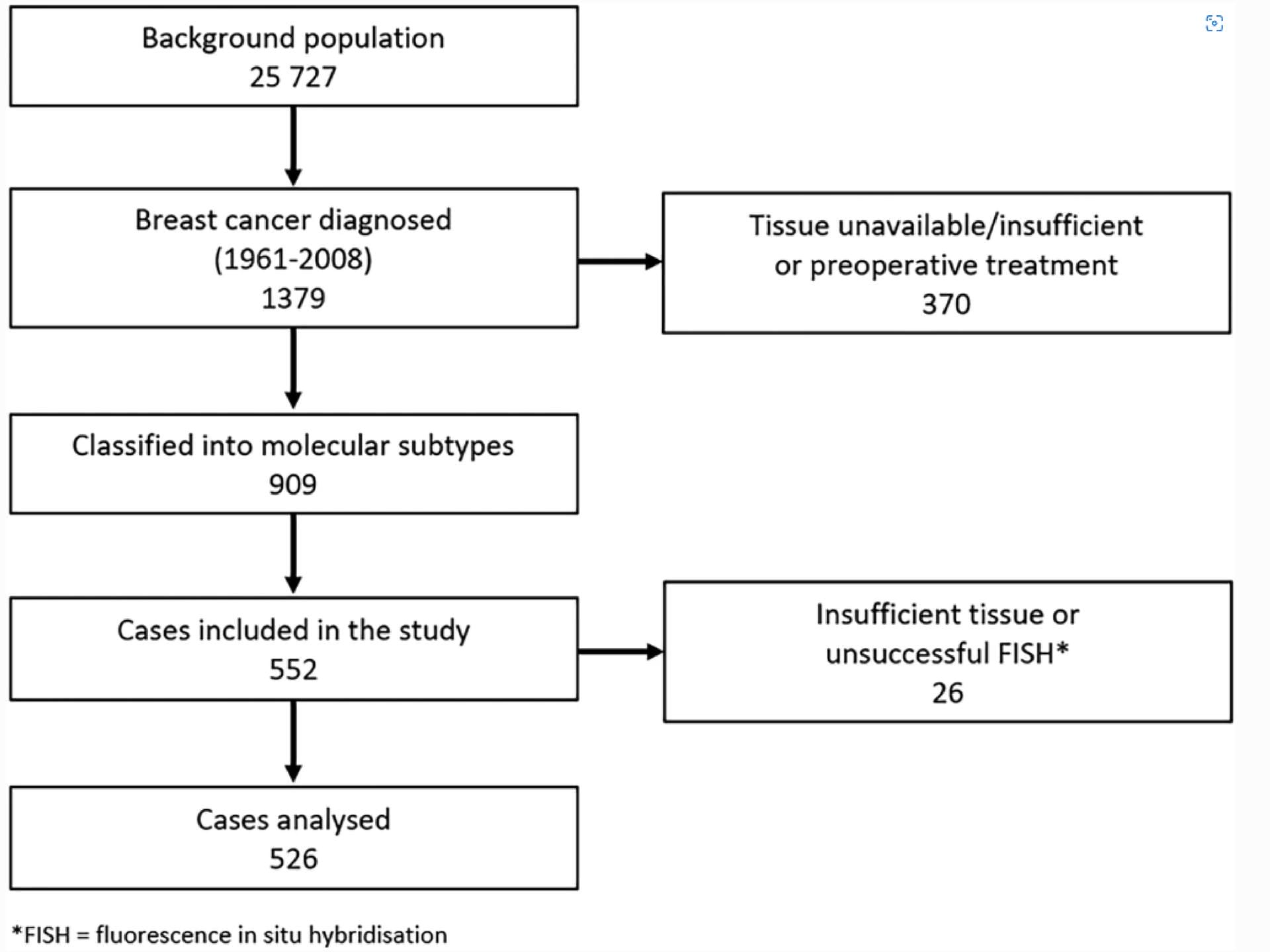
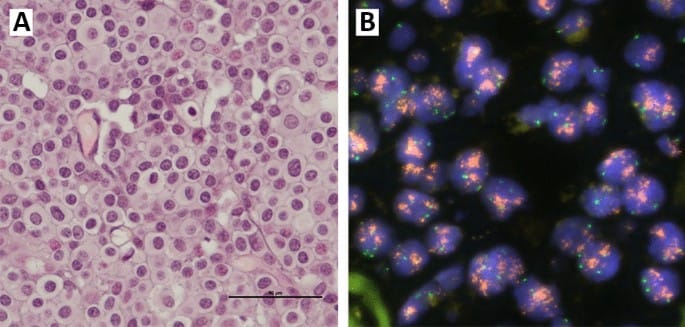
This study used fluorescence in situ hybridization (FISH) to target CCND1 and the chromosome 11 centromere (using chromosome 11 enumeration probe, CEP11). For each tissue sample, CCND1 and CEP11 copy number were counted in 20 non-overlapping, well-preserved tumor cell nuclei. The authors then calculated mean CCND1 and mean CEP11 copy number per tumor cell nucleus for each of 526 cases. An increase in CCND1 copy number (mean ≥4) was found in 16.6% of cases. The increase in CCND1 copy number was not accompanied by an increase in CEP11 copy number, which suggests gene amplification of CCND1 in these cases. The study authors also reported good correspondence between CCND1 copy number in the primary tumors and CCND1 copy number in the corresponding lymph node metastases.
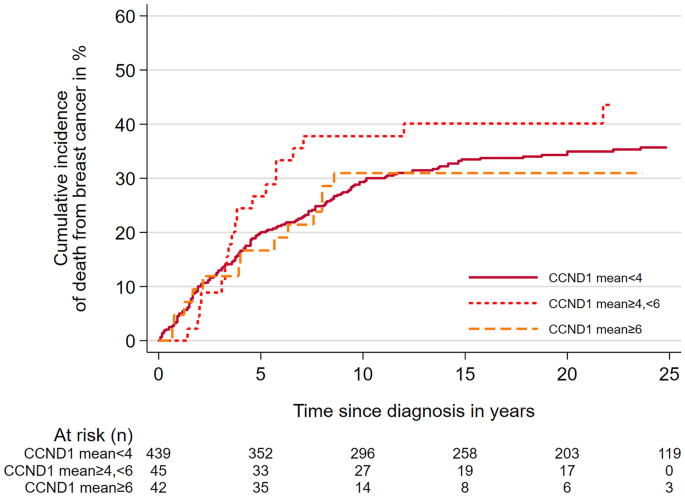
The authors observed high mean CCND1 copy number (≥6) in all molecular subtypes. The highest proportion of cases with high mean CCND1 copy number was seen among Luminal B (HER2-) cases; this represented a statistically significant association (p = 0.002). There was also a significant association between high mean CCND1 copy number (≥6) and histopathological grade 3 tumors (p=0.007). Finally, there were statistically significant associations between CCND1 copy number and measures of proliferation; cases with increased CCND1 copy number were also more likely to be Ki67 high (p < 0.0001) and have higher mitotic counts (p = 0.006). Despite these results, there was no statistically significant association between high CCND1 copy number and poor prognosis.
Since it has been suggested that CCND1 cooperates with other genes in major oncogenic pathways, the authors also looked for an association between amplification of CCND1 and amplification of two genes located at 8p12, FGFR1 and ZNF703. Some breast cancers exhibiting CCND1 amplification also show amplification of these genes. Although sample sizes for these comparisons were generally small, the authors noted that when mean copy number of two or more of these genes was high (≥6), the tissue sample tended to have a higher histopathological grade, be Ki67 high, and have high mitotic counts, confirming that these three genes are associated with highly proliferative breast cancer. Due to the fact that multiple molecular events may lead to gene amplification, coupled with the complexity of these multigene interactions, further studies will be needed to clarify the role of CCND1 as a prognostic biomarker.
